- 1. Why sRNAPrimerDB?
- 2. When to use sRNAPrimerDB?
- 3. How can I design primers or probes for small non-coding RNA?
- 4. What the definition of advanced parameter?
- 5. How can I search primers or probes in sRNAPrimerDB?
- 6. How to perform your experimental?
- 7. How to submit your validated primers or probes?
- 8. How to download primers or probes sequences?
- 9. How do I report problems or suggestions?
- 10. One-step RT-PCR vs. two-step RT-PCR?
- 11. End-point RT-PCR vs. real-time RT-PCR?
- 12. Troubleshooting?
- 13. Policies for sRNAPrimerDB usage
sRNAPrimerDB is a comprehensive web primer or probe design tool specifically for small non-coding RNAs (sncRNAs), such as microRNA (miRNA, 20-25 nts), PIWI-interacting RNA (piRNA, 24-32 nts), short interfering RNA (siRNA, 20-25 nts), etc. sRNAPrimerDB allow users to design several types of primers including generic or specific reverse transcription primers (Linear poly(T) RT primers, Linear S-poly(T) RT primers, stem-loop RT primers, stem-loop poly(A) RT primers, stem-loop poly(T) RT primers, stem-loop S-poly(T) RT primers), specific PCR forward and reverse primer pairs, double hairpin probe, hybridization oligos for nine detection methods (Table 1), such as A. Linear poly(A) tailed RT-PCR, B. Linear S-poly(A) tailed RT-PCR, C. Stem-loop RT-PCR, D. Stem-loop poly(U) tailed RT-PCR, E. Stem-loop poly(A) tailed RT-PCR, F. Stem-loop S-poly(A) tailed RT-PCR, G. One-step real time RT-PCR, H. Double-Stem-loop LH-PCR, I. Northern blot or ISH. In addition, sRNAPrimerDB is also a primers bank, which contains tens of thousands of primer pairs for detecting miRNAs and piRNAs.
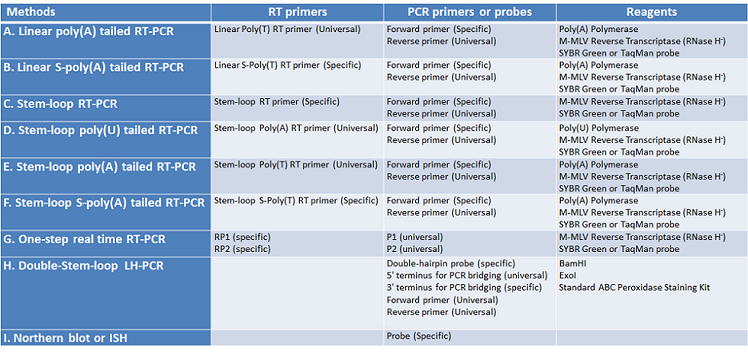
Table 1.Primer design characteristics of different detection methods.
To study small ncRNA expression, firstly, the appropriate experimental methods needs to be determined (method A to I). Secondly, high-quality RNA must be extracted in quantities sufficient for subsequent electrophoresis separation of RNA and hybridization, or cDNA library construction. Thirdly, prepare primers or probes, reagents and equipment according to different experiments. Here, sRNAPrimerDB can be used to design primers or probes for nine experimental methods(Table 1). Final, the hybridization experiment or RT-qPCR reaction can be performed. The expression level of small ncRNAs can be obtained by analyzing the experimental data (Fig 1).
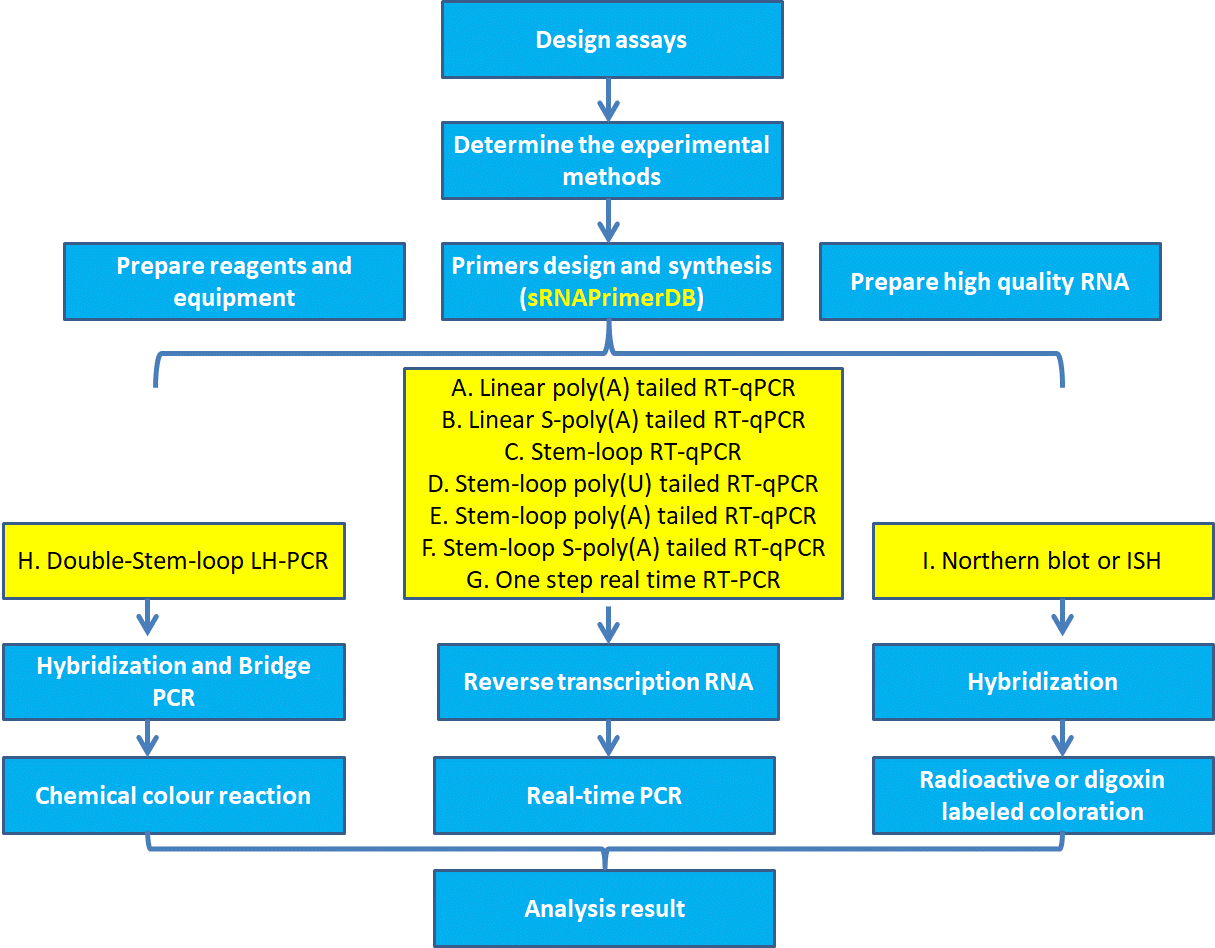
Fig 1. Flowchat. Once the primers, probes, reagents have been designed, received, and prepared, it is advisable to test them before assaying experimental samples.
A. Go to the Design web page.
B. Select experimental method, for example "A. Linear poly(A) tailed RT-PCR" in the drop down "Design” box.
C. Paste small ncRNA sequence in the "Small ncRNA sequences (fasta format)" box.
D. Select the appropriate source organism in "Background DNA" box that is likely to contain the target sequence.
E. Click the "Run" button, this program will run according to the default parameters to design primers or probes for small ncRNAs.
F. If you want to adjust the best parameters, you can click the "Advanced Parameters" options. Then you can set your own RT primer sequence, Specific Length (bp), Gap Length (bp), Optimal Tm (℃), Tm cutoff (℃), Tm difference (℃), Primer size (bp), Primer GC (%), Concentration of monovalent cations (usually KCl), Concentration of dNTPs, Concentration of divalent cations (usually MgCl2) and Annealing oligo concentration (described in more detail below).
G.The program will list out various types of primer sequences. For qRT-PCR assay, sRNAPrimerDB provides a RT primer and 5 pairs of candidate PCR primer pairs. For hybridization assay, sRNAPrimerDB provides a probe for Northern-blot /ISH.
H. Double check to make sure there are no hairpins or dimmers from the bottom right corner of the program.
I. Click the "Download all records" link to download all results.
J. Order your primer/probe from a company that offers custom oligonucleotides. Make sure that you label them in a way that you will know which one is which. Cut and paste the sequence from your record when you can to avoid typo. When you receive the primers from the company make a 0.5 mM stock solution in PBS or DNase/RNase free water (50 nmol in 0.1 ml). Keep it in the freezer.
RT primer: This refers to the reverse transcription primer. For different experimental methods, which use different type of RT primers, for example, linear poly(T) RT primers, linear S-poly(T) RT primers, stem-loop RT primers, stem-loop poly(A) RT primers, stem-loop poly(T) RT primers, stem-loop S-poly(T) RT primers.
Specific Length: This refers to the length of the portion of your RT primer that is binding to the 3’ of small ncRNA, the default length is 6 base pairs.
Gap Length: This refers to the gap between specific RT primers and forward primers relative to small ncRNA, the default length is 2 base pairs.
Optimal Tm: This refers to the optimal primer melting temperature (Tm). Primers with melting temperatures in the range of 55-60 ℃ generally produce the best results.
Tm cutoff: This refers to the minimum Tm value required for the binding stability between primer and its binding sites. The default value is 30 ℃.
Tm difference: This refers to the maximum allowable melting temperature difference between forward and reverse primers. The default value is 5 ℃.
Primer size: This refers to the primer length, it is generally accepted that the optimal length of PCR primers is 18-25 bp.
Primer GC: This refers to the GC content allowable in your primers, computed as a percentage over the entire primer length. If this value doesn't matter, then just set the range from 0 to 100.
A. Go to the Primer Bank web page.
B. Select small ncRNA species, for example "miRNA" in the drop down "Primer Bank" box.
C. Select "Search by experimental method".
D. There are several ways to search for primers or probes: miRBase miRNA ID or you can search your mature miRNA sequence against the sRNAPrimerDB sequence database.
A. Go to the Protocol web page.
B. Select experimental method, for example "A. Linear poly(A) tailed RT-PCR" in the drop down "Protocol" box.
C.The experiment is carried out according to the recommended protocol. You may need to modify this protocol if you use different reagents or instruments for real-time PCR.
A. Go to the Submit web page.
B. Select experimental method, for example "A. Linear poly(A) tailed RT-PCR" in the drop down "Submit" box.
C. Different primer pairs or probes should be submitted according to different experimental methods. Please ensure that the primer pairs or probes have been verified by experiments, and please submit relevant articles for verification.
A. Go to the Downloads web page.
B. Select small ncRNA species, for example "miRNA" in the drop down "Search" box.
C. Different primer pairs or probe data sets can be downloaded according to different species and different experimental methods.
Please tell us what you think about our web site and our primers or probe in primer submission page. If you provide us with your email, we will be able to reach you in case we have any questions.
The quantification of small ncRNA using RT-PCR can be achieved as either a one-step or a two-step reaction. The difference between the two approaches lies in the number of tubes used when performing the procedure. In the one-step approach, the entire reaction from cDNA synthesis to PCR amplification occurs in a single tube. For instance, One-step real time RT-PCR. On the other hand, the two-step reaction requires that the reverse transcriptase reaction and PCR amplification be performed in separate tubes.
The measurement approaches of end-point RT-PCR requires the detection of small ncRNA gene expression levels by the use of fluorescent dyes like ethidium bromide. The emergence of novel fluorescent DNA labeling techniques in the past few years have enabled the analysis and detection of PCR products in real-time and has consequently led to the widespread adoption of real-time RT-PCR for the analysis of small ncRNA gene expression. Currently, there are two different fluorescent DNA probes available for the real-time RT-PCR detection of PCR products: SYBR Green and TaqMan probe. All of these probes allow the detection of PCR products by generating a fluorescent signal. While the SYBR Green dye emits its fluorescent signal simply by binding to the double-stranded DNA in solution, the TaqMan probes generation of fluorescence depends on Förster Resonance Energy Transfer (FRET) coupling of the dye molecule and a quencher moiety to the oligonucleotide substrates.
Here we listed a few major causes for real-time PCR failures. Please read the sRNAPrimerDB manuscript for more details.
Little or no PCR product. Poor quality of PCR templates, primers, or reagents may lead to PCR failures. First, please include appropriate controls to eliminate these possibilities. Some small ncRNAs are expressed only in certain tissues. Please first read literature to make sure your small ncRNAs are included in the cDNA templates. In our experience, this is the most likely cause for negative PCR results. If you are sure the small ncRNAs are expressed, then try lowering the annealing temperature (to Tm -5 °C) and increasing the annealing time to ensure sufficient primer annealing. If you still could not see any PCR band, it could mean the primer pair in use is the problem. Although great care has been given to design sRNAPrimerDB primers, the success rate is not 100%. Less than 20% of the primers may have design problems (see our paper for detailed discussion). In this case, please try a different primer pair or even different methods for the same small ncRNAs. If you have any questions please feel free to contact us.
Poor PCR amplification efficiency. The accuracy of real-time PCR is highly dependent on PCR efficiency. A reasonable efficiency should be at least 80%. Poor primer quality is the leading cause for poor PCR efficiency. In this case, the PCR amplification curve usually reaches plateau early and the final fluorescence intensity is significantly lower than that of most other PCRs. This problem may be solved with re-designed and synthesized primers.
Primer dimer. Primer dimer may be occasionally observed if the small ncRNAs expression level is very low. If this is the case, increasing the template amount may help eliminate the primer dimer formation.
Normalization control. Selection of reference genes for normalization of small ncRNAs expression by RT-qPCR. For example, U6 or 5s rRNA.
Multiple bands on gel or multiple peaks in the melting curve. Agarose gel electrophoresis or melting curve analysis may not always reliably measure PCR specificity. For short amplicons ( < 150 bp) very weak (and fussy) bands migrating ahead of the major specific bands are sometimes observed on agarose gel. These weak bands are super-structured or single-stranded version of the specific amplicons in equilibrium state and therefore should be considered specific. Although gel electrophoresis or melting curve analysis alone may not be 100% reliable, the combination of both can always reveal PCR specificity in our experience.
Non-specific amplicons. Non-specific amplicons, identified by both gel electrophoresis and melting curve analysis, give misleading real-time PCR result. To avoid this problem, please make sure to perform hot-start PCR and use at least 60C annealing temperature. We noticed not all hot-start Taq polymerases are equally efficient at suppressing polymerase activity during sample setup. The SYBR Green PCR master mix described here always gives us satisfactory results. If the non-specific amplicon is persistent, you have to choose a different primer pair for the small ncRNA of interest. You are also encouraged to report bad primers to Shengsong Xie (ssxie@mail.hzau.edu.cn).
How to cite sRNAPrimerDB?
You can cite the following papers describing the work done for the sRNAPrimerDB database. sRNAPrimerDB IDs should also accompany the cited primer pairs.
Xie et al., sRNAPrimerDB: A comprehensive primer design and search web service for small non-coding RNAs. 2018. In prepare.
Can I make a link to sRNAPrimerDB?
You are welcome to make a static link to sRNAPrimerDB on your web site. sRNAPrimerD is designed to provide free PCR primers or probes for all researchers. You may freely use these primers or probes in your research with appropriate acknowledgements. However, you must get written permission from us if you intend to use our data to set up a similar primer database to provide services to third-party users or if you intend to include more than 500 primer pairs in your commercial projects.